WHAT IS WHOLE EXOME SEQUENCING?
Navigating the complexities of genetic and rare conditions from diagnosis to treatment can be challenging, especially when answers feel out of reach. Determining the genetic cause of a patient’s condition is often crucial to effective medical management, targeted treatment, and informed genetic counseling. But when should you consider WES, and how can it transform the diagnostic journey?
Whole Exome Sequencing (WES) is a diagnostic tool that analyzes the protein-coding regions of the genome, which represent the majority of disease-causing genetic variants. This approach provides crucial insights into genetic conditions that might be missed by standard genetic testing, enabling earlier and more accurate diagnoses.
Exons are the short, functionally important sequences of DNA which represent the regions in genes that are translated into proteins; a gene can contain multiple exons. WES is a next-generation sequencing technique that sequences the exome (the entire set of exons) to find an underlying genetic cause (etiology) relating to the patient’s medical issues in a single test. 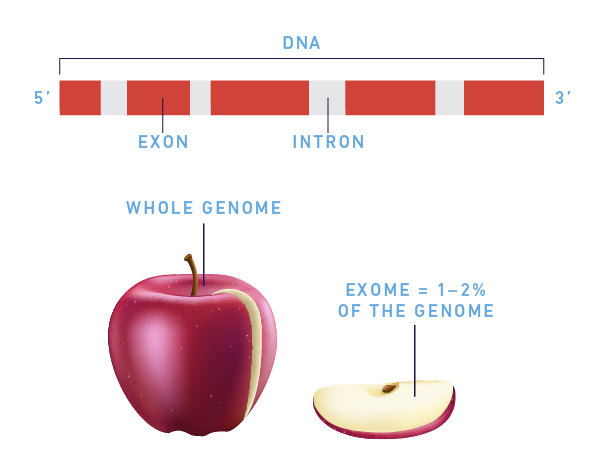
Figure 1: Understanding WES
While the exome only makes up about 1 – 2% of the genome, it is estimated to contain 85% of disease-causing mutations. By sequencing these regions, WES provides an in-depth look at genetic variants that may explain a patient’s clinical symptoms, identify genes implicated in genetic diseases, and assess recurrence risk.
WES can detect single nucleotide variants (SNVs), small insertions and deletions (indels), and larger copy number variations (CNVs) within the protein-coding regions of the genome, offering a thorough examination of the genetic landscape. It also has the ability to identify specific structural variants (SVs), further enhancing its diagnostic capabilities. Compared to targeted testing panels and standard cytogenetic tests, these advantages increase the chance for a diagnosis, accurate prognosis, and changes in medical management.
WES in Real World
WES is valuable for pediatric patients with conditions such as multiple congenital anomalies, neurodevelopmental disorders, and epilepsy where genetic etiology is suspected. Patients with multiple disorders often present with atypical phenotypes which leave physicians struggling to make a definitive diagnosis. Dual diagnoses can occur in pediatric patients, particularly those with conditions like epilepsy, multiple congenital anomalies, or unexplained symptoms. Research shows that 2% to 7% of patients with epilepsy have a dual diagnosis1. For patients in need of urgent answers, Rapid WES can offer quick results within days, enabling early intervention and improving outcomes, especially for conditions that contribute to morbidity and mortality.
For example, in a child with seizures, overgrowth, hemimegaloencephaly, cerebellar vermis hypoplasia, a single left palmar crease along with other dysmorphic features and worsening respiratory failure – the diagnostic path was far from straightforward. Concern for a somatic mosaic overgrowth condition led the genetics team to order Rapid Trio WES.
Rapid Trio WES revealed two separate diagnoses:
Familial Focal Epilepsy: A pathogenic variant in the NPRL3 gene was identified, inherited from the child’s father. This variant is associated with autosomal dominant familial focal epilepsy with variable foci, type 3.
Trisomy 21: An additional diagnosis of Down syndrome was made, which was confirmed through Chromosomal Microarray Analysis. This result explained the child’s dysmorphic features.
Impact: The dual diagnosis provided clarity in a complex situation, offering not only answers to the patient’s symptoms but also valuable insights for the family’s health management. WES testing offered a clear diagnosis, preventing a lengthy journey of uncertainty, and highlighting the benefits of early and comprehensive genetic testing.
- Guided Treatment: The care team implemented targeted therapies for familial focal epilepsy.
- Informed Family Planning: This result allows at-risk paternal relatives to undergo genetic testing and informs recurrence risks for the family. Additionally, the NPRL3 variant has variable penetrance, meaning it may or may not cause symptoms in individuals with the variant.
- Comprehensive Care: Guided by the American Academy of Pediatrics’ health supervision guidelines for children with Down syndrome, the family could be referred for specialty care and supportive therapies.
Identifying the root cause of a genetic condition, particularly early in the diagnostic journey, has significant implications for clinical management, enabling providers to make informed decisions regarding treatment, surveillance, and prognosis.
Early genetic testing not only enables the initiation of more targeted therapies but also empowers healthcare providers to implement more effective surveillance and monitoring strategies. Ultimately, this leads to better clinical outcomes and a higher quality of life for patients and their families. Additionally, WES can aid in risk assessment and family planning.
Supporting Research and Evidence
- In a 2024 study published in Genetics in Medicine, pathogenic or likely pathogenic variants were identified in 41.8% of 246 patients with neurodevelopmental disorders (NDDs). This resulted in 62 different genetic diagnoses, driving critical changes in clinical management including cascade testing (30.6%), family counseling (22.2%), medication adjustments (13.9%), clinical trial referrals (2.8%), medical surveillance (30.6%), and specialty referrals (69.4%). These findings underscore the value of genetic testing in patients with NDDs, where pathogenic variants are found in up to 50% of patients2.
- According to a study published in BMC Neurology of 101 patients diagnosed with moderate to severe ASD, 29.7% (30/101) of patients received a diagnosis through WES, detecting: SNVs in 27 patients and CNVs in 3 patients. Notably, WES captured all CNVs that CMA testing identified3.
- A study published in JAMA of over 400 patients with epilepsy who received a genetic diagnosis revealed that approximately 1 in 2 patients had changes in clinical management; in many, these changes were associated with reduction or elimination of seizures4.
Guidelines and Recommendations
Several professional societies have addressed the use of WES in clinical care. The American College of Medical Genetics and Genomics (ACMG) recommends WES as a first-tier test for pediatric patients with developmental delay, intellectual disability, and congenital anomalies. Endorsed by the American Epilepsy Society, the National Society of Genetic Counselors recommends WES for all individuals with unexplained epilepsy. The American Academy of Pediatrics (AAP) and the American Association of Neuromuscular & Electrodiagnostic Medicine (AANEM) both support the use of WES to achieve genetic diagnoses.
Advanced WES Solutions at Baylor Genetics
The diagnostic potential of WES is closely linked to the quality and rigor of the testing process. While WES provides comprehensive insights into an individual’s genetic makeup, not all laboratories offer the same robust features and capabilities to ensure optimal clinical utility.
Selecting the right laboratory partner is crucial to ensure the accurate detection of genetic variants and, when combined with RNA sequencing, achieve the most precise diagnoses for complex conditions. At Baylor Genetics, we offer:
- RNA Sequencing: Our enhanced WES includes complimentary RNAseq, an additional tool to aid in the interpretation of high-suspicion variants. Combining WES with RNA sequencing offers even greater potential to uncover genetic answers.
- Rapid Testing for NICU/PICU: Baylor Genetics’ Rapid WES returns written results in as few as 5 days for rapid testing and in 3 weeks for standard.
- Family Testing Options: Trio testing (patient and both parents) improves variant interpretation and helps clarify inheritance patterns.
- Flexible Sample Type Options: Flexible sample options, including blood, saliva, and tissue, ensure testing can continue even when standard samples are challenging to collect, making testing more accessible and tailored to patient needs.
- Insurance Coverage: Did you know? Major insurers including UnitedHealthcare (UHC) and Aetna cover WES for pediatric neurological conditions, with broad coverage available through most commercial payors and State Medicaid programs when clinical criteria are met. With extensive coverage in place, Baylor Genetics is committed to making these advanced tests accessible to all your patients.
Hope Through WES: Empowering Better Outcomes
In the complex genetic and rare disease journey, the power of comprehensive genetic testing goes beyond finding answers; it transforms lives by enabling earlier, more precise interventions.
WES serves as diagnostic partner to your clinical expertise, shedding light on the genetic causes of complex conditions and supporting informed clinical decision-making. WES has proven invaluable in diagnosing conditions ranging from epilepsy, autism spectrum disorder, and neurodevelopmental disorders to rare syndromes involving multiple organ systems. Its application continues to grow, offering hope to families who have spent years searching for answers.
As we continue to push the boundaries of genomic medicine, Baylor Genetics remains committed to empowering patients, families, and healthcare providers with the insights needed to transform complex conditions into actionable, effective care. Together, we’re shaping a future where no patient is left without answers, and every family can navigate their diagnostic journey with confidence and hope.
We invite you to explore our comprehensive suite of WES offerings to see how we can assist you in improving patient care.
References:
- Garcia, Aixa Gonzalez et al. P179: Never two late: A dual diagnosis of Noonan syndrome and hypophosphatasia in an adolescent patient with unusual presentation. 2023. Genetics in Medicine Open, Volume 1, Issue 1, 100208
- Besterman AD, Adams DJ, et al. Genomics-informed neuropsychiatric care for neurodevelopmental disorders: Results from a multidisciplinary clinic. 2024. Genet Med.
- Sheth, Frenny & Shah, Jhanvi & Jain, et al. Comparative yield of molecular diagnostic algorithms for autism spectrum disorder diagnosis in India: evidence supporting whole exome sequencing as first tier test. 2023. BMC Neurology.
- McKnight D, Morales A, Hatchell KE, et al. 2022 Genetic Testing to Inform Epilepsy Treatment Management from an International Study of Clinical Practice. JAMA Neurol. 79(12):1267–1276.

